Experiment ran for 2 weeks after each week once on novebmer 12th and once on november 19th we took tissue samples of the gills of one mussel from each treatment group.
Experimental set up
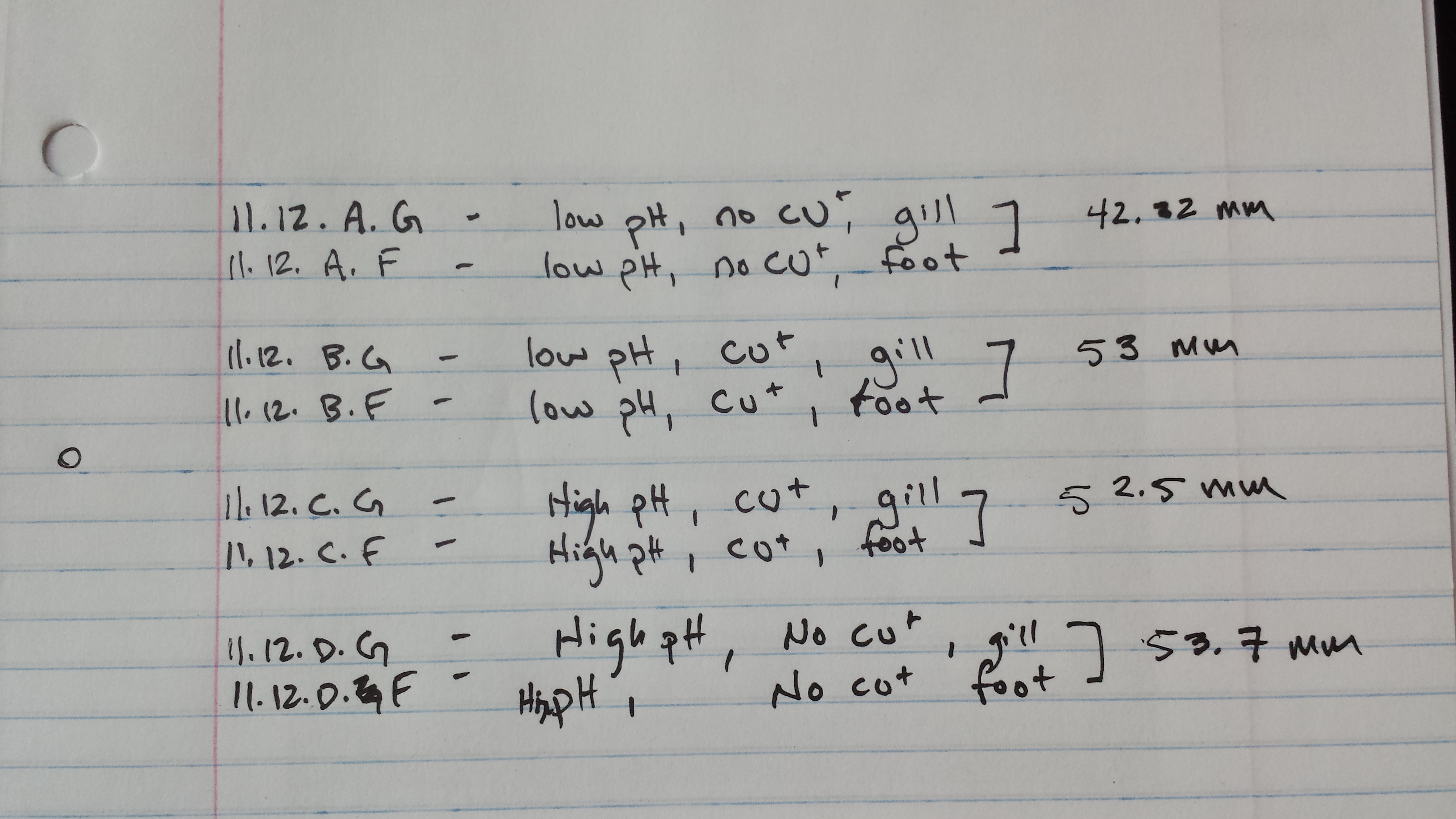 LAB 6
LAB 6
we took initial samples of tissue from two different mussles to use as a baseline in the rest of our experiment.Lab 5
Today in lab as a group we discussed what we would be doing for our projects.
As a group we got approval for our project.
LAB 4
Objectives
- Run extracted total protein from previous labs on SDS-PAGE Gel/Western Blot
- Run conventional PCR samples on an agarose gel
- Download qPCR data and discuss analysis
ELECTROPHORESIS PROCEDURE
- Place gel in gel box and fill with 1x TAE buffer (to fully cover wells)
- Remove combs from wells
- Load 7uL 100bp ladder in far left lane
- Load 20uL of your PCR sample into the gel (retain the remaining vol at -20ºC)
- Run gel at ~ 100V for ~ 40min
- Visualize the gel on the UV transilluminator
- Mine was in well 4
Protein Extraction and Analysis Part 2
- Have water boiling
- In a fresh, 1.5mL SCREW CAP tube add 15uL of your protein stock and 15uL of 2X Reducing Sample Buffer. Return your protein stock to the box in the -20C freezer labeled protein samples.
- Mix sample by flicking. Briefly centrifuge (10s) to pool liquid in bottom of tube.
- Boil sample for 5 mins.
- While sample is boiling, observe assembly of gel box and gels. Rinse gel wells thoroughly as demonstrated.
- When sample is finished boiling, immediately centrifuge for 1min. to pool liquid.
- Slowly load your entire sample into the appropriate well using a gel loading tip.
- Put lid on gel box and plug electrodes into appropriate receptacles on the power supply.
- Turn power supply on and set voltage to 150V. Run for 45mins. CHECK YOUR AGAROSE GEL RESULTS. MAKE SURE EVERYTHING IS SET UP FOR WESTERN BLOT.
- Turn off power supply and disconnect gel box from power supply.
- Remove lid from gel box.
- Disengage the tension wedge.
- Remove gel from gel box.
- Carefully crack open cassette to expose gel.
- Trim wells at top of gel.
- Notch a designated corner of the gel to help you remember the correct orientation of the gel (i.e. which is the top/bottom of the gel, which is the right/left side(s) of the gel)
- Proceed to Western Blotting protocol.
WesternBreeze Chromogenic Western Blot Immunodetection
Western Blot Protocol
This is a lengthy protocol with many incubation steps. There will be one gel for the entire class. As a class, you should assign yourselves steps so that everyone can participate and so that we don't waste time. When it is not your turn to attend to the gel, you can do protein and RNA extractions for your project.
- Soak the filter paper, membrane and gel in Tris-Glycine Transfer Buffer for 15 minutes.
- Assemble the blotting sandwich in the semi-dry blotting appartus:
- Anode (+++)
- filter paper
- membrane
- gel
- filter paper
- cathode (---)
- Transfer the blot for 30 minutes at 20V
- Remove the gel from the sandwich and rinse off adhering pieces of gel with transfer buffer.
- Wash membrane 2 times, for 5 minutes each, with 20 mL of pure water.
- Put the membrane in the plastic box and add 10 mL of Blocking Solution. Cover and incubate overnight on a rotary shaker set at 1 revolution/second.
- Your TA will do the rest of the steps. After class tomorrow you can come and see your results.
- Decant liquid.
- Rinse the membrane with 20 mL of water for 5 minutes, then decant. Repeat.
- Incubate the membrane in 10 mL of Primary Antibody Solution. Decant the solution.
- Rinse the membrane with 20 mL of Antibody Wash for 5 minutes, then decant. Repeat 3 times.
- Incubate the membrane in 10 mL of Secondary Antibody Solution for 30 minutes. Decant.
- Wash the membrane for 5 minutes with 20 mL of Antibody wash, then decant. Repeat 3 times.
- Rinse the membrane with 20 mL of pure water for 2 minutes, then decant. Repeat twice.
- Incubate the membrane in 5 mL of Chromogenic Substrate until a purple band appears. This will occur between 1-60 minutes after adding the Chromogenic Substrate.
- Dry the membrane on a clean piece of filter paper to the open air.
LAB 3
- Previously done: Reverse transcribe RNA to cDNA
- Perform qPCR on your cDNA samples
- Run qPCR samples on agarose gel
- Mock-up Experiment to prepare for the initiation of experiments
- Design and order primers
Supplies and Equipment:
- Micropipettes (1-1000 μl)
- Sterile filter pipette tips (1-1000 μl)
- Tip waste jar
- PCR tubes (0.5 ml; thin walled)
- RNA samples (student provided)
- M-MLV reverse transcriptase
- M-MLV 5X reaction buffer
- Oligo dT
- dNTPs
- Nuclease Free water
- thermal cycler
- microfuge tube racks
- PCR tube racks
- ice buckets
- Kimwipes
- Lab coat
- Safety glasses
- gloves
Procedure background
A reverse transcriptase, also known as RNA-dependent DNA polymerase, is a DNA polymerase enzyme that transcribed single-stranded RNA into double-stranded DNA. It also helps in the formation of a double helix DNA once the RNA has been reverse transcribed into a single strand cDNA. Normal transcription involves the synthesis of RNA from DNA; hence, reverse transcription is the reverse of this. The resulting cDNA is more stable than RNA (which degrades quickly) can be used for downstream applications such as measuring gene expression. Partial nucleotide sequences of cDNAs are often obtained as expressed sequence tags. (This text has been adapted from Wikipedia)
REVERSE TRANSCRIPTION PROTOCOL
- Mix your stock RNA sample by inverting tube several times.
- In a 0.5 ml PCR tube labeled with your initials and “cDNA” combine the following:
- 5 μl of YOUR total RNA (extracted and quantified in lab)
- 1 μl of oligo dT
- 4 μl of nuclease free H2O
- Incubate the mixture for 5 min at 70°C on the thermocycler then immediately transfer to ice. Briefly centrifuge you tube and the add the following:
- 5 μl of M-MLV 5X Reaction Buffer
- 5 ul of dNTPs
- 1 μl of M-MLV RT
- 4 μl of nuclease free H2O
- Incubate the mixture for 60 min at 42°C and then heat inactivate at 70°C for 3 min on the thermocycler.
- Spin down the sample in a desk top centrifuge.
- Store on ice or at -20°C
Quantitative PCR
Supplies and Equipment:
- PCR Plates (white); optically clear caps
- 1.5 ml microfuge tubes (RNAse free)
- Nuclease Free water
- filter tips
- Opticon thermal cycler
- kim wipes
- 2x Immomix Master Mix
- SYTO-13 Dye
- microfuge tube racks
- ice buckets
- timers
- cDNA samples (student provided)
qPCR PROCEDURE
You will run each template (cDNA) in duplicate in addition to two negative controls (no template) - calculate how many reactions this will be!
1. Prepare master mix: Prepare enough master mix for your number of reactions +1 to ensure sufficient volume recovery.
Master Mix: 62.5 uL
upstream primer 2.5uL
downstream primer2.5uL
ultra pure water52.5uL
2. Add mastermix to wells of a white PCR plate
3. Thaw cDNA samples.
4. Add 1uL cDNA template to each reaction.
5. Add 1uL of ultra pure water to the negative control wells.
6. Cap the wells securely.
7. If necessary, spin the strips to collect volume in the bottom of the wells.
8. Ensure the lids are clean and place strips on ice. (I like to wipe the lids with a clean kimwipe)
9. Load the plate, verify the PCR conditions and start the run (this will be done by your TA).
PCR conditions:
1. 95°C for 10 minutes
2. 95°C for 15s
3. 55 °C for 15 s
4. 72°C for 15 s (+ plate read)
5. Return to step 2 39 more times
6. 95°C for 10s
7. Melt curve from 65°C to 95°C, at 0.5°C for 5s (+plate read)
Protein Extraction and Analysis Part 1
Supplies and Reagents
- micropipettes (1-1000uL)
- sterile filter pipette tips (1-1000uL)
- sterile (RNase free) 1.5mL microcentrifuge tubes
- sterile 2 mL screw cap microcentrifuge tubes
- sterile disposable pestles
- spectrophotometer
- cuvettes for spectrophotometer
- microcentrifuge (refrigerated) or in fridge
- ice buckets
- gloves
- Kim wipes
- lab pens
- safety glasses
- CelLytic MT Cell Lysis Reagent (with Protease Inhibitor Cocktail added)
- Coomassie Protein Assay Reagent
- DI water
Procedure Background
This section provides an explanation of the methods being used and provieds some essential background information.
- You will isolate cellular protein from whole tissue using CelLytic MT. This is a proprietary reagent that contains a mixture of salts and detergents to effectively disrupt lipid membranes, lyse cells, and buffer the cellular proteins at the appropriate pH. This solution will also be supplemented with a cocktail of protease inhibitors to minimize the impact of the numerous proteases that are ubiquitous within all cells.
- After extraction, you will determine the concentration of proteins in your sample. Due to the high variability of protein structures, these molecules do not uniformly absorb light at any specific wavelength like nucleic acids. We will use the Bradford Assay to determine the concentration of proteins in your sample. This is a colorimetric assay that uses a reagent (Coomassie Blue) that interacts with proteins. When the reagent is mixed with a solution containing proteins, the solution will turn different intensities of blue depending on the amount of proteins present in the sample. This blue dye absorbs at 595nm and the absorbance can be directly correlated to a specific amount of protein present in the sample when compared to a standard curve.
- A standard curve is created by conducting the assay with a set of proteins of known concentration (standards). A dilution series is created from these standards to span the maximum possible range of detection for the assay. This dilution series is then processed according to the assay protocol and measurements are taken. The measurements resulting from these standards are then plotted on a graph and a best fit line is created using these data points. From this best fit line, an equation can be obtained for the slope of the line (y = mx+b). This equation allows you to now put in any value you obtain from an experimental sample and determine the concentration of proteins in that sample.
- Due to the dynamic nature of the reaction taking place in the Bradford Assay (and virtually all assays), a standard curve should be created using the exact same reagents and equipment that will be used for the experimental samples. This helps to account for variation between different pieces of equipment as well as slight differences between reagent lots. Ideally, a new standard curve should be made each time a set of experimental samples are being assayed to ensure the best accuracy. For the sake of time, a standard curve has already been determined.
- Mixing - The Bradford assay works best when samples are mixed well. Invert tubes frequently during incubations, and immediately before measuring absorbance to ensure accurate absorbance readings.
PROTEIN EXTRACTION PROTOCOL
- Record the weight of your tissue that has been denoted on the tube.
- Label the snap cap tube containing your tissue sample with your initials and the date using a lab marker.
- Add 500 ul of CellLytic MT solution to the 1.5mL snap cap tube containing your cut piece of frozen tissue.
- Homogenize the tissue with a sterile disposable pestle.
- Close the tube and invert the tube several times.
- Please find a few other people at or near this same stage and form a group for this step. Spin the tube in a refrigerated microfuge for 10 mins at max speed.
- While spinning, label a fresh tube with the word "Protein", source organism/tissue, your initials, and today's date.
- Carefully transfer supernatant (the clearish liquid on top) to labeled tube and store tube on ice.
PROTEIN QUANTIFICATION PROTOCOL
- Lable a fresh 2 mL screw cap tube withe the word "Protein", BA (for Bradford Assay), your initials, and today's date.
- Dilute an aliquot of your protein sample 1:2 by pipetting 15uL of your protein sample into the 2 mL screw cap tube and the pipetting 15uL of DI water. Mix well by pipetting. Note: this dilution step is performed to ensure the sample absorbance falls within the range of the standard curve
- In a second 2 mL tube pipette 30uL of DI water (this tube will serve as your blank). Label tube as 'blank'
- To both tubes add 1.5mL of Bradford reagent. Tip: Pippet 1000ul of reagent into each tube and then pipet another 500 ul of reagent into each tube for a total of 1500u ul or 1.5 mL.
- Invert the tubes several times and then incubate at room temperatire (RT) for 10mins.
- Mix the 'blank' via pippeting and transfer to a 1000ul to a plastic, disposable cuvette.
- Zero the spectrophotometer using your blank sample. Be sure to wipe the cuvette with a KimWipe first as any fingerprints or smudges can alter the reading.
- Mix the 'sample' via pippeting and transfer 1000 ul to a plastic, disposable cuvette
- Measure the absorbance at 595nm and record the value. Be sure to wipe the cuvette with a KimWipe first as any fingerprints or smudges can alter the reading.
- Remove the cuvette from the spectrophotometer. Using a P1000 set to 1000 ul, carefully pipette the solution in the cuvette up and down a couple of times to mix.
- Measure the absorbance at 595nm and record the value a second time.
- Average the two absorbance values you recorded.
- Back-calculate your protein concentration using the standard curve below. Hint: Use the equation on the graph provided. The relationship between absorbance and concentration is linear and defined by the equation y=mx+b. You have the average absorbance of your sample, x, and you want to calculate the concentration, y. Don't forget to account for the dilution in step 2!
- Give your protein sample to the TA for storage at -20ºC.
| x |
----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
LAB 2
- You will isolate RNA from whole tissue using TriReagent. TriReagent allows for separation of RNA from other cellular components, including DNA. There are three primary components of TriReagent that allow this to happen. The first is guanidine isothiocyanate which is a potent protein denaturant, the second is phenol, and the third is pH.
- Guanidine isothiocyanate denatures proteins, such as the highly abundant histones that coat DNA. Even more importantly, RNases are denatured. This denaturing action allows for better access of phenol (an organic solvent) to cellular proteins and improves its ability to keep the proteins insoluble. The pH of TriReagent is acidic. The low pH keeps DNA out of solution while RNA remains soluble.
- After homogenizing/lysing your tissue in TriReagent, chloroform (another organic solvent) will be added to your sample to allow for separation of the phenol and insoluble cellular components (DNA, proteins) from soluble cellular components (RNA). This will result in three distinct layers: the organic phase (the bottom portion), the interphase (layer of cell debris) and the aqueous phase (the top portion). The aqueous phase (the RNA) can then be easily isolated.
- The RNA can be precipitated and washed to remove residual phenol and salt carryover. Then the RNA can be resuspended in a suitable solution and quantitated.
- RNA is quantitated using a spectrophotometer and measuring the absorbance of your RNA sample at 260nm (A260). The concentration of your sample is calculated with the following equation:[RNA] = 40ug/mL x A260 x Dilution Factor. The spectrophotometer we will be using calculates the concentration for us using this equation and the absorbance measurement.
- The purity of your sample can be assessed using the ratio of A260 to A230 and to A280. Various substances will absorb at 230nm, which will indicate carryover of phenol, ethanol or high salt in your sample. Proteins generally absorb light at 280nm. For clean RNA, A260/A280 should range between 1.8-2.0. The A260/A230 should range between 1.5-2.0 for clean RNA. For more information on interpreting these ratio see the Nanodrop user manual.
For a fun introduction to quantification using the Nanodrop please watch this video.
RNA EXTRACTION PROTOCOL
Continued from Lab 1
1. Turn on heating block to 55°C.
2. Incubate your homogenized tissue sample (from Lab 1) tube at room temperature (RT) for 5 mins.
3. In the fume hood, add 200uL of chloroform to your sample and close the tube. NOTE: Due to the high volatility of chloroform, pipetting needs to be done carefully and quickly. Have your tube open and close to the container of chloroform before drawing and chloroform into your pipette tip.
4. Vortex vigorously for 30s. You are vortexing correctly if the solution becomes a milky emulsion.
5. Incubate tube at RT for 5 mins.
6. Spin tube in refrigerated microfuge for 15 mins. @ max speed.
7. Gently remove tube from microfuge. Be sure not to disturb the tube.
8. Slowly and carefully transfer most of the aqueous phase (the top, clear portion) to a fresh microfuge tube. Do NOT transfer ANY of the interphase (the white, cell debris between the aqueous and organic phase).
9. Close the tube containing the organic and interphase and properly dispose of the liquid inside the tube as well as the tube itself at the end of the lab.
10. Add 500uL isopropanol to the new tube containing your RNA and close the tube.
11. Mix by inverting the tube numerous times until the solution appears uniform. Pay particular attention to the appearance of the solution along the edge of the tube. If mixed properly, it should no longer appear viscous/"lumpy".
12. Incubate at RT for 10 mins.
13. Spin in refrigerated microfuge at max speed for 8 mins. When placing your tube in the microfuge position the tube hinge pointing up, away from the center of the microfuge.
14. A small, white pellet (RNA and salts) should be present. If not, do not fret an continue with the procedure.
15. Remove supernatant.
16. Add 1mL of 75% EtOH to pellet. Close tube and vortex briefly to dislodge pellet from the side of the tube. If the pellet does not become dislodged, that is OK.
17. Spin in refrigerated microfuge at 7500g for 5mins.
18. Carefully remove supernatant. Pellet may be very loose. Make sure not to remove pellet!
19. Briefly spin tube (~15s) to pool residual EtOH.
20. Using a small pipette tip (P10 or P20 tips), remove remaining EtOH.
21. Leave tube open and allow pellet to dry at RT for no more than 5mins.
22. Resuspend pellet in 100uL of 0.1%DEPC-H2O by pipetting up and down until pellet is dissolved.
23. Incubated tube at 55C for 5mins. to help solubilize RNA.
24. Remove tube from heat, flick a few times to mix and place sample on ice. This will be your stock RNA sample.
25. Quantitate RNA yield using Nanodrop spectrophotometer.
RNA QUANTIFICATION
NOTE: Always keep your RNA samples on ice!
1. Pipette 2µL of 0.1%DEPC-H20 onto the Nanodrop pedestal and lower the arm.
2. Click "Blank", to zero the instrument. NOTE: steps 1 and 2 only need to be done once for the whole class.
3. Pipette 2µL of your RNA sample onto the Nanodrop pedestal and lower the arm
4. Click "Measure". Record your RNA concentration (ng/µL), A260/280 ratio and A260/230 ratio. NOTE: The Nanodrop uses the Beer-Lambert Law to calculate RNA concentration for you. See Lab 1 notes on RNA extraction for more information on the calculation and how to evaluate RNA purity using A260/280 and A260/A230 ratios.
6. Raise the arm and wipe off you sample with a KimWipe
7. Clearly label your stock RNA sample with the word "RNA", source organism/tissue, your initials, today's date and the concentration in ug/uL.
8. Give your samples to the TA for storage at -80C.
- NOTE: we moved onto lab 3 and did not quantify the data called for in this lab
- Mix your stock RNA sample by inverting tube several times.
- In a 0.5 ml PCR tube labeled with your initials and “cDNA” combine the following:
- 5 μl of YOUR total RNA (extracted and quantified in lab)
- 1 μl of oligo dT
- 4 μl of nuclease free H2O
- Incubate the mixture for 5 min at 70°C on the thermocycler then immediately transfer to ice. Briefly centrifuge you tube and the add the following:
- 5 μl of M-MLV 5X Reaction Buffer
- 5 ul of dNTPs
- 1 μl of M-MLV RT
- 4 μl of nuclease free H2O
- Incubate the mixture for 60 min at 42°C and then heat inactivate at 70°C for 3 min on the thermocycler.
- Spin down the sample in a desk top centrifuge.
- Store on ice or at -20°C
Lab 1
RNA Extractions part 1
Supplies and reagents
- Micropipettes (1- 1000uL)
- sterile filter pipette tips (1- 1000uL)
- sterile (RNase free) 1.5 mL microcentrifuge tubes
- sterile disposeable pestiles
- vortex
- ice buckets
- cloves
- lab pens
- safety glasses
- triReagent
- I isolated RNA from whole tissue using TriReagent. this allows for separation of RNA from other cellular components, including DNA.the triReagent is made up of guanidine isothicoyanate which is a potent protein denaturant, the second is phenol, and the third pH
- These substansas denatures proteins, such as highly abundant histones that coat DNA. Even more importantly, RNases are denatured. This denaturing action allows for better access of thenol (an organic solvent) to cellular proteins and improves its ability to keep the proteins insoluble. The PH of TriReagent is acidic. The low pH keeps DNA out of solution while RNA ramains soluble
- After homogenizing/ lysing your tissue in TriReagent, chloroform
- RNA Isolation Protocal
- Label the snap cap tube containing your tissue sample your initials and the data using a lab marker. Keep the sample stored on ice until you are ready for homogenization
- add 500uL of TriReagent to
- the 1.5 mL snap cap tube containing your tissue. store on ice
- carefully homogenize the tissue using a disposable pestle. If the tissue. store on ice
- carefully homogenize the tissue using a disposable pestle. If the tisue is difficult to homogenize carefully close the tube tightly and briefly cortex the sample
- after the sample is completely homogenized,and close the tube tightly
- vortex vigorously for 15 s
- stop her for lab one and after labeling store tissue sample in a freezer.
- Using a sterile pastle, homogenize your tissue sample in 0.5mL of DNazol in a 1.5mL sterile microfuge tube. After the tissue is homogenized, add 0.5mL more of DNazol and mix well
- let your sample incubate for 5 miniutes at room temperature
- spin your sample at 10,000x g (room temp) for 10 miniutes
- With my sample I had to spin it twice
- transfer your supernatant to a new labeled tube
- add 0.5 mL of 100% ethanol to your sample
- mix your sample by inverting your tube 5-8 times
- store your sample at room temperature for 1 minute
- your DNA should form a cloudy precipitate. Remove the DNA and put in a new tube using your pipette
- let your sample sit at room temp for 1 miniute and remove the rest of the lysate (liqhid that is not DNA)
- wash your DNA with 1 mL of 75% ethanol: Pipette the ethanol into your DNA tube, invert 6 times, and let sit for 1 minute. Remove the ethanol from the tube and repeat.
- If there is ethanol left at the bottom of your tube after the second wash, remove with a small pipette
- add 300 uL of 0.1% DEPC water to yourDNA and pipette up and down multiple times to dissolve
- bring your DNA sample to the Nanodrop to quantify
the results where
Sample 6
ABS 8.757
lambda 230
A-260 4.240
A-280 2.198
260/ 280 1.93
260/230 .48
212.0 ng/ul